Roberto Bonasio, Ph.D.
Professor Of Cell And Developmental Biology

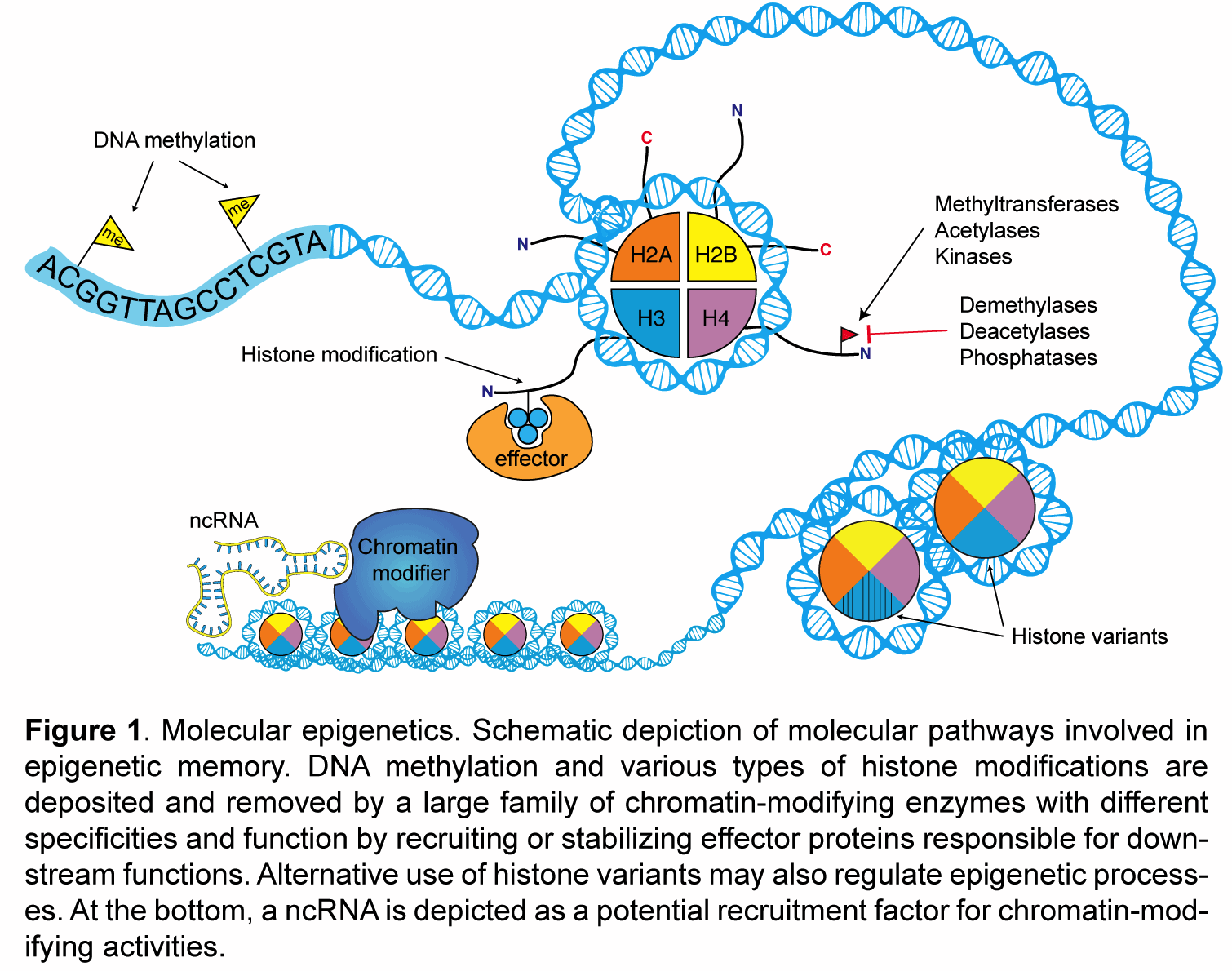
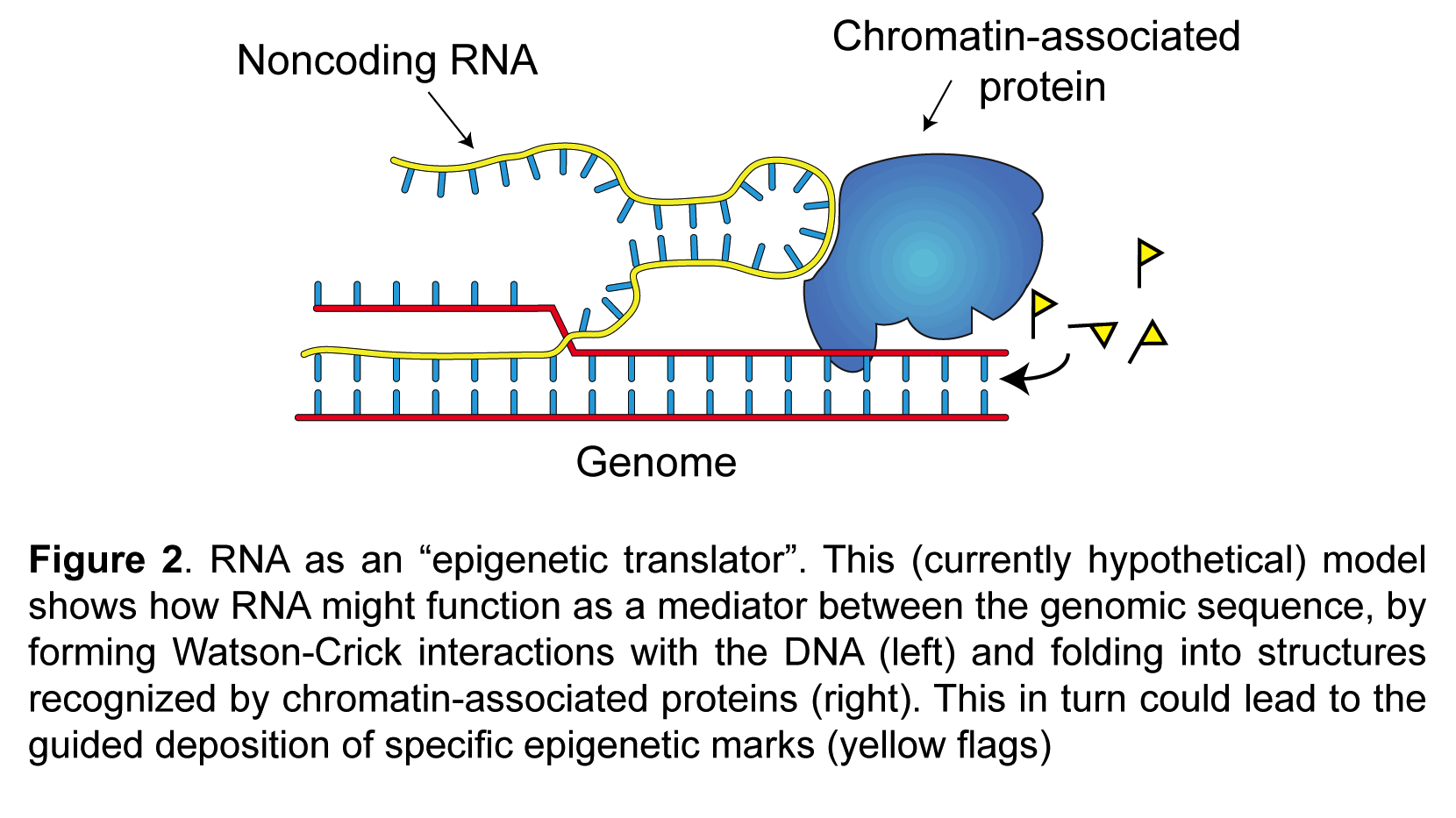
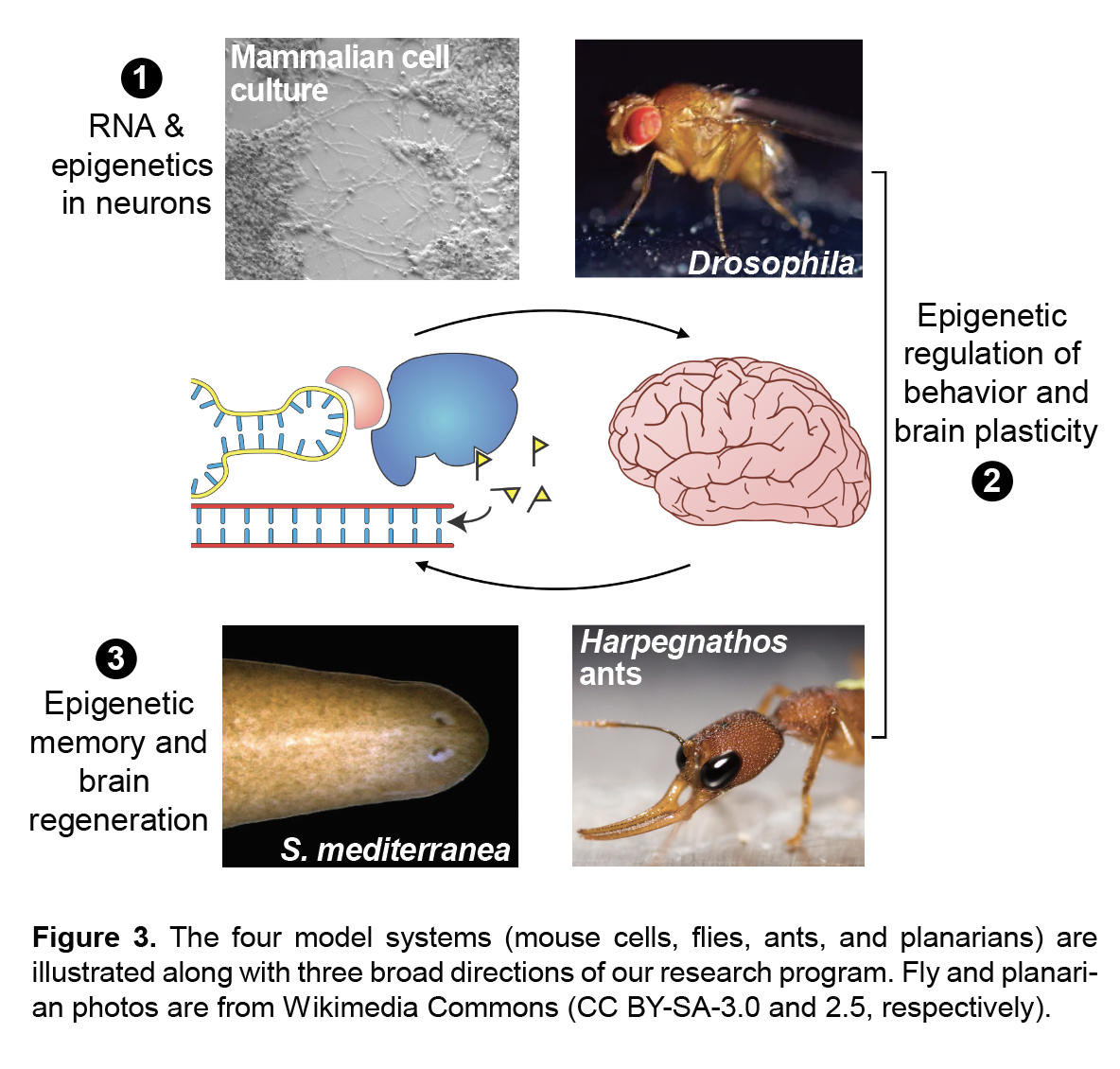
My laboratory studies the molecular mechanisms of epigenetic memory, which are key to a number of biological processes, including embryonic development, cancer, stem cell pluripotency, and brain function.
- Molecular mechanisms of epigenetic memory
- Noncoding RNAs
- Chromatin biochemistry
- Genes and behavior

- Laurea (Biotechnology) University Of Milan, 2000.
- PhD (Immunology) Harvard Medical School, 2006.